Looking for Help with Assignments?
-RUCats has 80 hours of tutoring available online and in-person. Check the tutoring tab in Canvas!
-Instructors and Lead TAs have a combined 10 hours of Office Hours, open to all sections. See times/locations here.
-Piazza (found in the Canvas sidebar) provides fast support from Course Staff and other Students. Be sure to search to see if someone has asked a similar question!
-If you need a computer to complete work on, iLab machines can be found in the CAVE (Hill 252) and surrounding rooms.
Forensic Analysis – 98 course points
The purpose of this assignment is to practice using Binary Search Trees to analyze DNA.
Start your assignment early! You need time to understand the assignment and to answer the many questions that will arise as you read the description and the code provided.
Refer to our Programming Assignments FAQ for instructions on how to install VSCode, how to use the command line and how to submit your assignments.
- See this video on how to import the project into VSCode and how to submit into Autolab.
The assignment has two components:
- Coding (95 points) submitted through Autolab.
- Reflection (3 points) submitted through a form.
- Submit the reflection AFTER you have completed the coding component.
- Be sure to sign in with your RU credentials! (netid@scarletmail.rutgers.edu)
- You cannot resubmit reflections but you can edit your responses before the deadline by clicking the Google Form link, signing in with their netid, and selecting “Edit your response”
Overview
You’ve been tasked with identifying who a DNA sequence may belong to based on an unknown person’s DNA sequence and a DNA database you have. A person’s DNA has pairs of chromosomes; we will focus on a single pair of the same chromosome to identify people whose samples we want to investigate further.
DNA is a molecule found in living organisms that carries our genetic information. It is made up of nucleotides, which are organic building blocks that consist of a base, sugar, and phosphate. Nucleotides contain one of four different bases – adenine (A), thymine (T), cytosine (C), and guanine (G). We will focus on combinations of these four bases in this assignment.
99.9% of our DNA is the same as other humans. Our goal is to focus on the 0.1% of DNA that varies between humans – these are sections of “noncoding DNA” that are in between genes and don’t provide instructions for making proteins. Short Tandem Repeats, or STRs, are in noncoding DNA and are short sets of bases that are repeated across a sequence.
In this assignment, you will work with simple DNA sequences with 2-5 STRs. We will identify the number of times a set of STRs is repeated within a sequence to find out who a pair of unknown DNA sequences may belong to in a DNA database.
Why BSTs? In a past semester of CS111, students had a DNA assignment that used an array of objects to store a DNA sequence and find hereditary relationships. When we have a large number of items, we can add/remove a single item more efficiently using a BST in O(log n) time than an array, which takes O(n) time. Do you understand why this is?
- This assignment differs significantly from the CS111 assignment: it focuses on BST operations to access and flag profiles rather than string and object operations to calculate STRs within profiles. We provide you with the STRs.
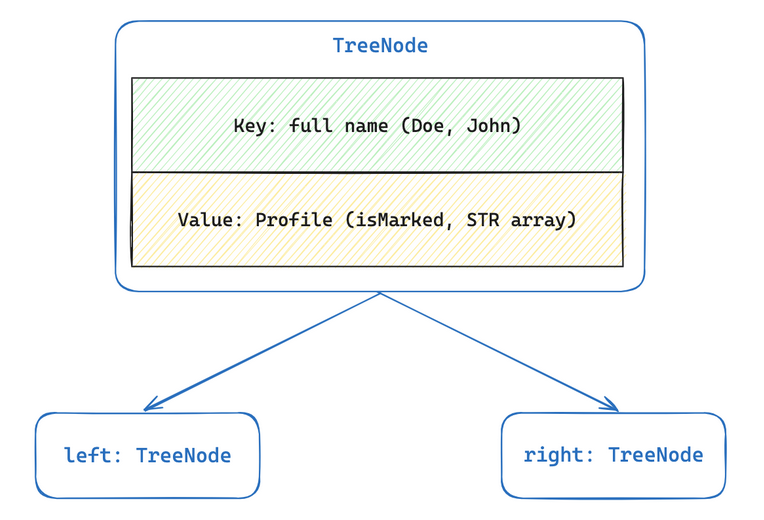
- Here’s a diagram that shows how we are using nodes to store (name, profile) pairs:
Implementation
Overview of files provided
- The Profile class allows you to create instances of Profiles; it stores information whether the profile is marked as a profile of interest as well as an array of STRs (ie. which STRs appear in a person’s DNA and how many times). Do not edit or submit to Autolab.
- The ForensicAnalysis class contains methods that manipulate BSTs in order to conduct STR analysis. Edit the empty methods with your solution, but DO NOT edit the provided ones or the method signatures of any method. This is the file you submit.
- The Driver class is used to test your methods interactively. Follow the prompts in the terminal to use the driver. Feel free to edit this file, as it is provided only to help you test your code. It is not submitted and it is not used to grade your code. Do not submit to Autolab.
- The TreeNode class represents a BST node; it contains a full name as its key, Profile as its values and TreeNodes “left” and “right” representing the left and right subtrees of the binary search tree. Getters and setters are provided. Do not edit or submit to Autolab.
- The Queue class is provided from the textbook; it contains enqueue and dequeue operations. Do not edit or submit to Autolab.
- StdIn and StdOut are libraries that handle input and output. Do not edit these classes.
- Multiple input files are included; they store data handling DNA profiles and the unknown chromosome parts. You are welcome to create your own input files, as they will not be submitted to Autolab.
ForesicAnalysis.java
- DO NOT add new import statements.
- DO NOT change any of the method’s signatures.
Methods to be implemented by you:
buildTree (provided)
This method reads a DNA database from an input file as well as two chromosomes belonging to an unknown person. It then calls the following methods to insert profiles into a BST rooted at treeRoot.
- This method reads the full name of a person (“Last, First” including the space in between, case-sensitive).
- This method calls createSingleProfile to read and return a single profile from the input file.
- Then, it calls insertPerson to create a TreeNode containing the person’s name and newly built profile and insert the profile into treeRoot.
Recall that:
- The reference variable treeRoot is the root of the BST.
- The BST node class is called TreeNode. Each TreeNode has a profile associated with it.
- The BST then holds all the (name, profile) pairs that are in the database.
Do not update this method, it has been implemented for you. IMPLEMENT the first two methods (createSingleProfile and insertPerson).
1. createSingleProfile
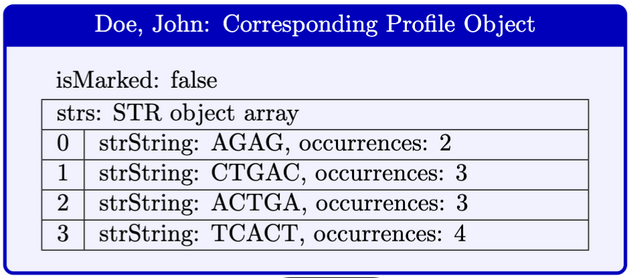
Given a single person’s data in an input file, read all information and return a new Profile containing the person’s information.
You have been provided with input files to test this method. Each input file is formatted as follows.
- One line containing a string with the person of interest’s FIRST sequence [DONE for you in buildTree]
- One line containing a string the person of interest’s SECOND sequence [DONE for you in buildTree]
- One line containing the number of people in the database, say p [DONE for you in buildTree]
- For each of p people:
- One line contains first name, last name, and number of STRs (say s), space separated. Names are read for you in buildTree!
- s lines containing an STR name and number of occurrences in that person’s DNA, space separated
To complete this method, do the following:
- For a single person in the database:
- Read the number of STRs (say s) associated with the person. You will create an array of STR objects of length s.
- For each STR:
- Read the STR name and number of occurrences.
- Create a new STR object, and add it to the next open space in the array.
- Create a Profile object using the data you just read, and return this profile.
Here is a diagram that illustrates a Profile object.
Use the StdIn library to read from a file:
- StdIn.setFile(filename) opens a file to be read. This is already done for you.
- StdIn.readInt() reads the next integer value from the opened file (whether the value is in the current line or in the next line).
- StdIn.readString() reads the next String value from the opened file.
Here is the expected output when testing createSingleProfile using the driver on input1.in. The driver calls the method on all people but does not update the unknown sequence instance variables. It also skips over names.
- We expect STRs to be in the SAME ORDER as the input files.

2. insertPerson
Given a full name (key) and Profile object (Value) passed through parameters, add a node containing this name profile to the BST rooted at treeRoot. Remember that treeRoot is a reference to the root node of the BST.
- Follow the BST insertion algorithm as discussed in class to insert Profiles into the BST.
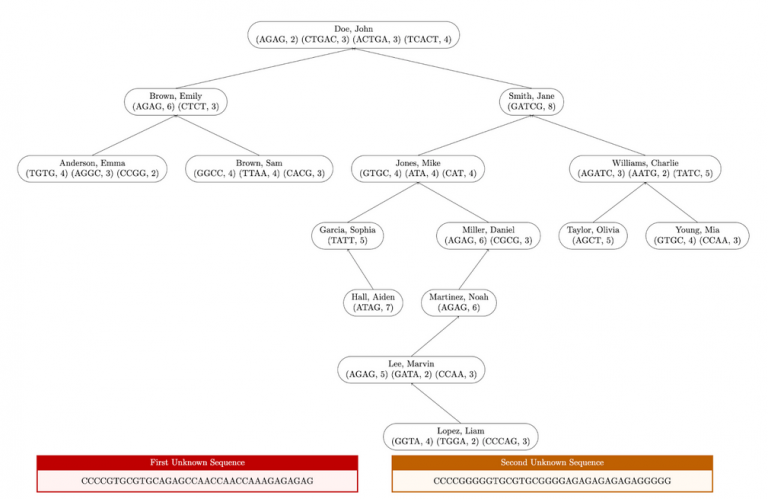
- Names are unique, nobody will share the same name.
- Follow BST ordering when inserting a profile. Use the compareTo method on the keys (full names) for comparisons. TreeNodes with names greater than a node go to the right of the node, otherwise they will go to the left.
Here is a diagram that illustrates the correct output for the entire structure in input1.in.
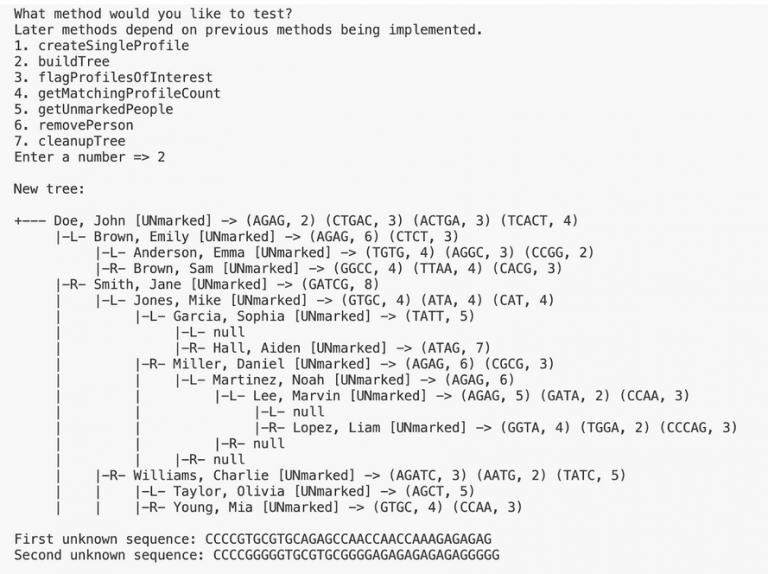
Here is the expected output when testing this method on input1.in. Test this method by calling buildTree in the driver, it requires createSingleProfile to be completed.
Early submission. Implement the first two methods for extra credit: createSingleProfile() and insertPerson().
numberOfOccurrences (provided)
This method takes in a sequence and STR. It returns the number of times an STR appears in the parameter sequence. This method is provided, do not edit it.
- You will call this method later in flagProfilesOfInterest.
3. flagProfilesOfInterest
This method marks profiles where for all STRs in a profile, most (at least half) of the STR occurrences in a profile match the occurrences of the same STR in the first and second unknown sequences combined. This is a simplified matching algorithm, and in reality the identification process is more complex.
- Traverse the BST rooted at treeRoot to investigate whether each profile should be marked.
- For a single profile, it is of interest if at least half of its STRs occur the same amount of times in both the profile, and the first+second sequence.
- We determine half of STRs by rounding UP to the nearest whole number if there is a remainder. For instance, if there are 3 STRs in a profile, half of 3 is 1.5, which rounds up to 2.
Hint: Use the provided numberOfOccurrences method to check the number of times an STR is repeated in a sequence.
In the following diagram, nodes with marked profiles are red and bold. Unmarked profiles are uncolored and unbolded. Profile STRs that appear in either unknown sequence are highlighted.
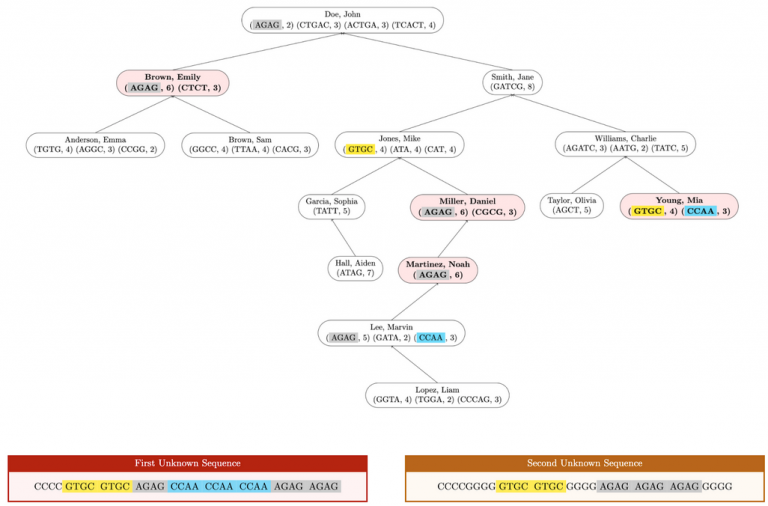
Here is the expected output when testing this method on input1.in. Test buildTree in the driver before testing this method:
4. getMatchingProfileCount
Find the number of profiles in the BST rooted at treeRoot whose marked status matches isOfInterest. Return this number. If there are no profiles that are marked according to the parameter isOfInterest, return 0.
- Recall that Profiles are values stored inside nodes. The Profile.java class contains a boolean instance variable isMarked and getters and setters. By default, all profiles are marked false (not profiles of interest).
- isMarked is a boolean value: true represents “marked” and false represents unmarked.
- Implement this method recursively. You are encouraged to use helper methods, just make them private.
In the following diagram, nodes with marked profiles are red and bold. Unmarked profiles are blue and unbolded.
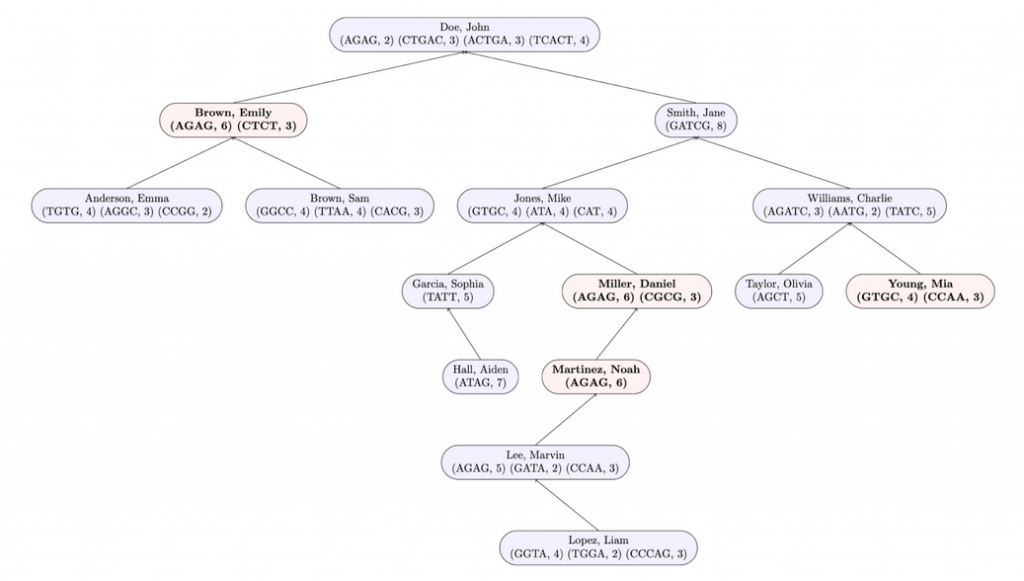
Here is the expected output when testing this method on input1.in. Test buildTree and flagProfilesOfInterest in that order in the driver before testing this method.
5. getUnmarkedPeople
This method uses a level-order traversal to populate and return an array of unmarked profiles in the BST rooted at treeRoot.
- Use the getMatchingProfileCount method you implemented earlier to get the array length. Create a new array of Strings using the number of unmarked profiles as its length.
- Use the provided Queue class to initialize a queue of TreeNodes. Until the queue is empty:
- For each node, investigate whether its profile is marked or not.
- If its profile is not marked, add the PERSON’S NAME (key) to the next available space in your resulting array.
- Enqueue the node’s children.
- To initialize a queue of TreeNodes, do: Queue<TreeNode> queue = new Queue<>();
- queue.enqueue(item) enqueues the parameter item into a queue.
- queue.dequeue() dequeues an item as discussed in class.
- queue.isEmpty() checks if the queue is empty.
- queue.enqueue(item) enqueues the parameter item into a queue.
- For each node, investigate whether its profile is marked or not.
- Return the resulting array.
Algorithm Example:
- The queue starts as empty. Start by enqueuing the root (at position 1).
- Dequeue the node at position 1, visit it, and enqueue its left and right children (positions 2 and 3).
- Repeat the same procedure on all other nodes until the queue is empty:
- Dequeue the node at position 2, visit it, and enqueue its children.
- Dequeue the node at position 3, visit it, and enqueue its children.
- Lastly, dequeue and visit positions 4 and 5. They are leaves.
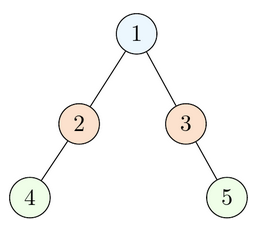
If the nodes at all these positions were marked, they would appear in the resulting array in ascending order of positions.
Input File Example: Unmarked profiles are in blue and bold and their corresponding index in the resulting array is next to their name. Marked profiles are in white.
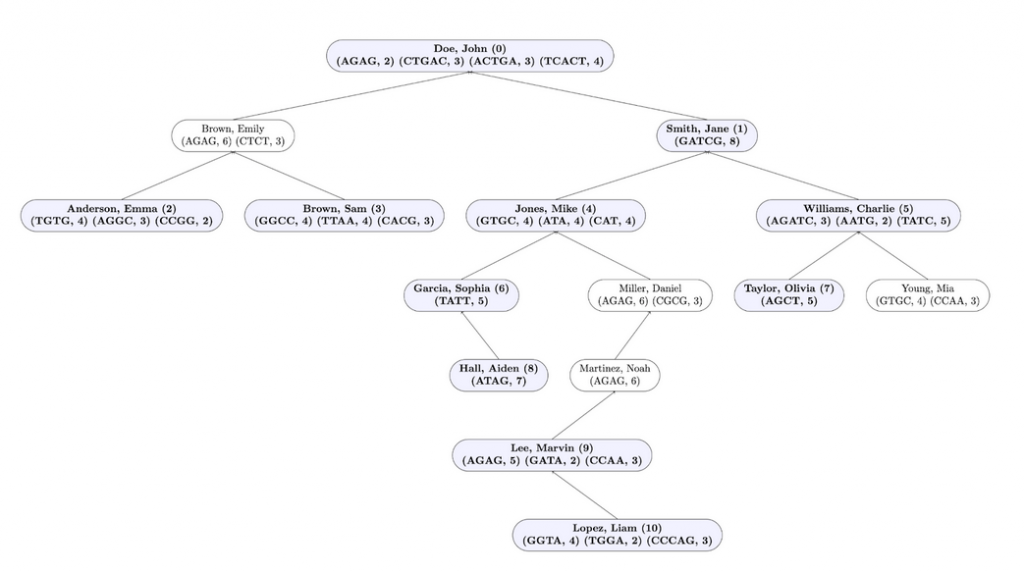
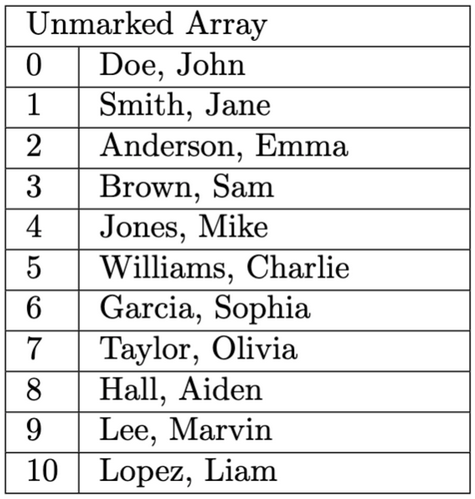
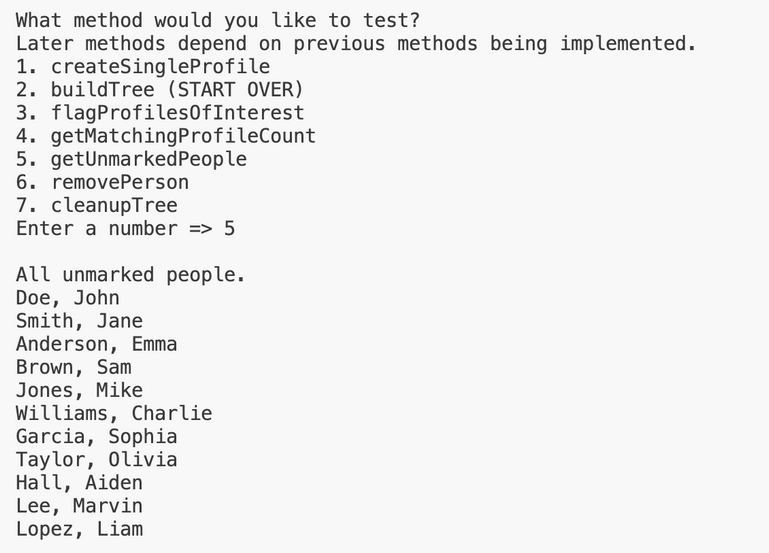
Here is the output for this method when testing this method on input1.in. Test buildTree and flagProfilesOfInterest in that order in the driver before testing this method, and IMPLEMENT getMatchingProfileCount before starting this method.
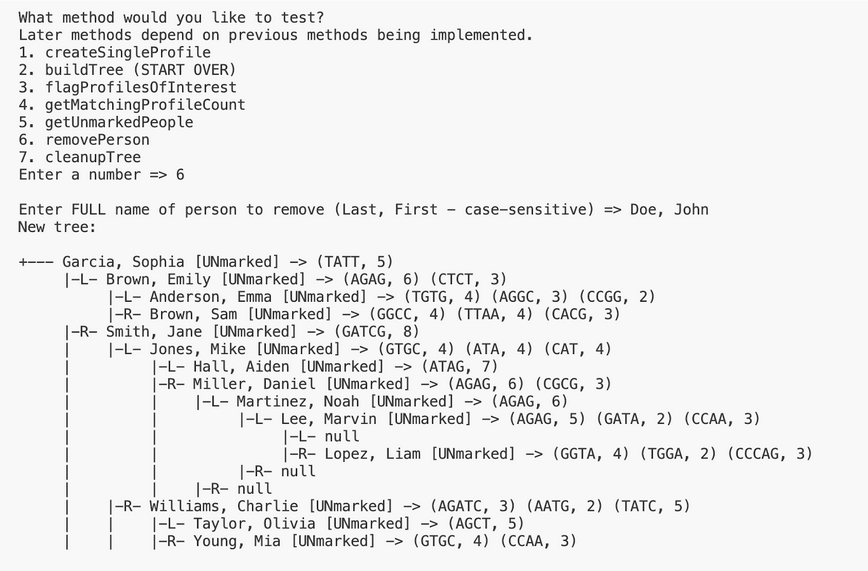
6. removePerson
This method removes a node whose name matches parameters. This is similar to the BST delete you have seen in class.
- Compare full names (“Last, First” including the comma and space) by using the compareTo method on keys as shown in class.
- If there are no left or right children, delete the node by setting the parent to null.
- If there is only one child, replace the parent link.
- If there are two children, find the inorder successor of the node, delete the minimum in its right subtree, and put the inorder successor in the node’s spot.
- Note: if the root of the BST changes treeRoot should be updated accordingly.
Here is the output for this method when testing this method on input1.in using “Doe, John”. Test buildTree in the driver before testing this method, and make sure all methods above are implemented before moving onto the next method. If you have tested other methods, start over.
7. cleanupTree
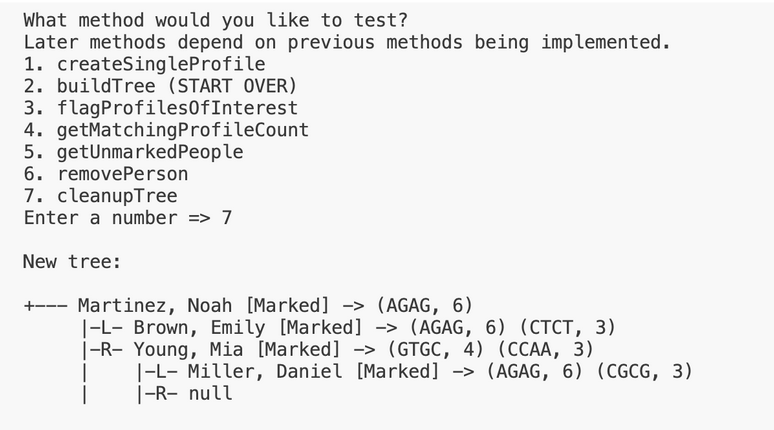
This method removes nodes containing unmarked profiles from the BST.
- Use the getUnmarkedPeople method to get an array of unmarked people.
- Use the removePerson method on each unmarked person, passing as parameters the full name of the person.
Here is the output for this method when testing this method on input1.in. Test buildTree and flagProfilesOfInterest in the driver in this order before testing this method, and make sure ALL methods are implemented. Start over if you have called removeProfile in the driver immediately before calling cleanupTree.
Implementation Notes
- YOU MAY only update the methods with the WRITE YOUR CODE HERE line.
- COMMENT all print statements you have written from ForensicAnalysis.java
- DO NOT add any instance variables to the ForensicAnalysis class.
- DO NOT add any public methods to the ForensicAnalysis class.
- DO NOT add/rename the project or package statements.
- DO NOT change the class ForensicAnalysis name.
- YOU MAY add private methods to the ForensicAnalysis class.
- YOU MAY use any of the libraries provided in the zip file.
- DO NOT use System.exit()
VSCode Extensions
You can install VSCode extension packs for Java. Take a look at this tutorial. We suggest:
Importing VSCode Project
- Download ForensicAnalysis.zip from Autolab Attachments.
- Unzip the file by double clicking.
- Open VSCode
- Import the folder to a workspace through File > Open
Executing and Debugging
- You can run your program through VSCode or you can use the Terminal to compile and execute. We suggest running through VSCode because it will give you the option to debug.
- How to debug your code
- If you choose the Terminal:
- first navigate to ForensicAnalysis directory/folder
- to compile: javac -d bin src/forensic/*.java
- to execute: java -cp bin forensic.Driver
- first navigate to ForensicAnalysis directory/folder
Before submission
COMMENT all printing statements you have written from ForensicAnalysis.java
Collaboration policy. Read our collaboration policy here.
Submitting the assignment. Submit ForensicAnalysis.java separately via the web submission system called Autolab. To do this, click the Assignments link from the course website; click the Submit link for that assignment.
Getting help
If anything is unclear, don’t hesitate to drop by office hours or post a question on Piazza.
- Find instructors office hours here
- Find tutors office hours on Canvas -> Tutoring
- Find head TAs office hours here
- In addition to office hours we have the CAVE (Collaborative Academic Versatile Environment), a community space staffed with lab assistants which are undergraduate students further along the CS major to answer questions.
Inspiration from a Nifty assignment by Brian Yu and David J. Malan.
By Kal Pandit

